TRANSLATIONAL GENOMICS OF CARDIAC, PULMONARY AND NEUROLOGICAL DISORDERS.
Every 4 minutes, a baby is born with a birth defect. Rapid advances in next-generation sequencing have identified candidate gene variants. However, many variants remain of unknown significance (VUS) and a huge burden on birth defect diagnosis. Clinicians cannot use VUS in decision-making and, therefore, cannot 1) assess patients’ risk of complications, 2) provide the best clinical therapy, and 3) provide recurrence risk counseling for parents. Our long-term goal is to improve clinical outcomes for patients with birth defects by increasing the rate of genetic diagnosis.
RESEARCH
VISION
To be at the forefront of the cutting-edge field of functional genomics, enabling scientific curiosity, excellence in research, and groundbreaking discoveries to improve the clinical outcomes for babies born with cardiac, pulmonary, or neurological birth defects.
RESEARCH APPROACH
We use in vivo frog and mouse models and stem cell-based 2D and 3D (organoid) models to functionally characterize VUS in birth defects. We employ an interdisciplinary approach combining CRISPR/Cas9 genome editing, super-resolution microscopy, mechanobiology, single-cell/nuclei transcriptomics, and computational modeling.
PROJECTS
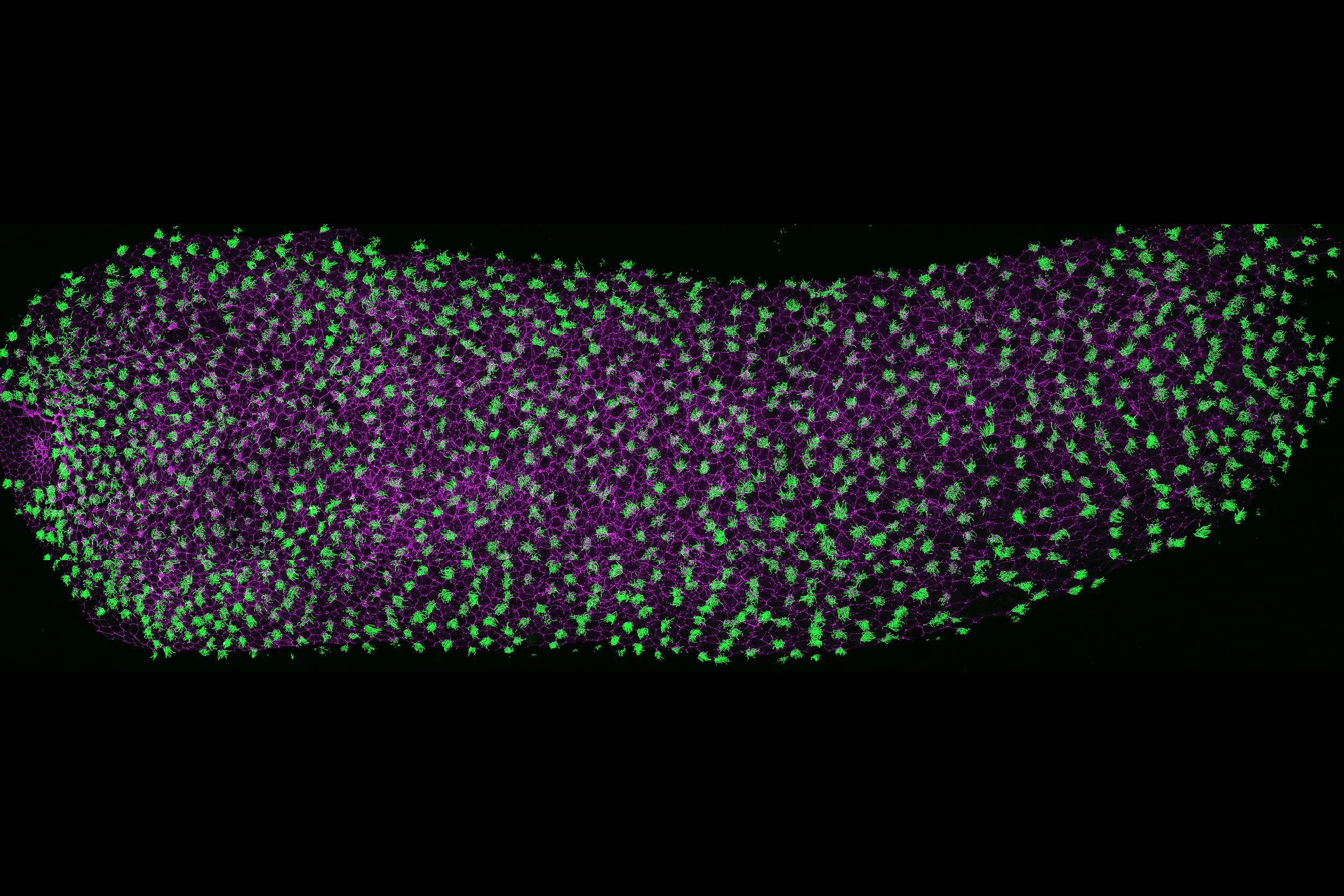
Discovering new genes and signaling pathways essential in cilia assembly and function in heart, lung, and brain development.
Cilia are microtubule-based organelles present on almost all the cells in the human body and serve multiple roles during development and homeostasis. Defects in the assembly or function of cilia can lead to an array of diseases, including congenital heart disease (cardiac), primary ciliary dyskinesia (pulmonary), and hydrocephalus and autism (neurological) disorders. We have projects focused on identifying novel genes and signaling pathways essential in cilia assembly and function in heart, lung, and brain development. Our research directly contributes to improving the depth of gene panels used for the genetic diagnosis of these birth defects.
Uncovering the mechanochemical gene regulatory networks (GRNs) in lung development and function.
Mechanical forces play a critical role in respiratory physiology. Our lungs continuously expand and compress as we breathe, exerting mechanical tension on the lung epithelium. This mechanical tension is essential for the differentiation of airway and lung epithelial cells (multiciliated cells, mucus-secreting cells, ionocytes, etc.). We have projects focused on uncovering mechanochemical GRNs essential for proper cell differentiation and maintaining epithelium integrity in the airway and lungs.
High-throughput translational genomics of pulmonary disorders.
Respiratory diseases (RDs) are caused by defects in the structure or function of motile cilia and multiciliated cells in the airway and lungs. These patients are at risk of recurrent respiratory infections, bronchiectasis, or even respiratory failure. About half of all these patients also develop cardiac defects. RDs are often challenging to diagnose due to the lack of a “gold standard” diagnostic tool and clinically variable presentation. Genetic testing has the potential to address these shortcomings. It can help identify patients who need functional assessment and improve the diagnostic process, thus impacting the need to diagnose patients at an early age. Next-generation sequencing (NGS) has identified about 60 genes, and about 1800 variants are classified as pathogenic. Yet, the rate of genetic diagnosis remains very low because an additional 10,000 variants remain of unknown significance (VUS) and a significant burden on RD diagnosis. We are addressing this shortcoming by developing and implementing high-throughput functional genomics models of frog embryos and human lung cell culture to classify variants robustly and at an unprecedented rate. Our research has an immediate positive global impact by increasing the rate of genetic diagnosis in RD patients in a clinically consequential time frame.
High-throughput translational genomics of congenital heart disease (CHD).
CHD is a leading cause of infant death. Nearly 1 in 100 babies are born with a heart defect, and about 25% have critical CHD. CHD remains a leading cause of infant death because phenotypically similar patients can have dramatically different outcomes. Therefore, we need clinical treatment driven by the genotype instead of a phenotype to treat these patients effectively. The NGS has identified candidate disease variants rapidly and cost-effectively. However, the yield of current clinical genetic testing is less than 30%, and determining the pathogenicity of the remaining candidate genes remains a major obstacle. Dr. Kulkarni has established a Bench-to-Bedside (B2B) CHD program at UVA Children’s to address this challenge. Our B2B project brings together experts in functional genomics (Dr. Kulkarni), genomic bioinformatics analysis (Dr. Ratan), genetic counseling (Mr. Thomas), pediatric cardiology (Dr. Peroutka and Dr. Haregu), and pediatric pulmonology (Dr. Garrod) to facilitate patient-driven research. Our research directly helps patients and families with aggressive management of clinical complications, including the risk of developing life-threatening conditions like respiratory problems and hydrocephalus. Further, our research enables recurrence risk counseling and reproductive decision-making for parents.
High-throughput translational genomics of neurological disorders.
NGS has identified hundreds of candidate genes and thousands of variants in neurological disorders like hydrocephalus and autism. However, their role in causing these disorders is not characterized. We address this unmet clinical need by functionally classifying these candidate genes and variants as pathogenic or benign to increase the diagnostic rates of these neurological disorders. Mouse is too slow and costly for the rapid functional classification of hundreds of variants. Therefore, we use frogs to functionally characterize these genes and variants in the context of brain development first. Analyssi in frogs is followed by mouse and human cell culture and organoid models for selective candidates. Our research has an immediate positive global impact by increasing the rate of genetic diagnosis in patients with hydrocephalus or autism.
MEET THE TEAM
-

Saurabh Kulkarni
Assistant Professor
-

Chris Courtney
Lab manager
-

Venkatraman Rao
Senior Postdoctoral Associate
-

Vani Narayan
Postdoctoral Associate
-

Vignesh Aravind
Postdoctoral Associate
-

Angelo Arrigo
Graduate Student
-

Victoria Hua
Undergraduate Researcher
-

Kranti Kaur
Undergraduate Researcher
-

Aarathi Manchikalapudi
Undergraduate Researcher
-

Krishna Girish Kumar
Undergraduate Researcher
RESOURCES
MICROSCOPY
NIKON AX-R confocal: This new state-of-the-art microscope is equipped with 8K Galvo and 2K resonant scanners, 4 solid-state laser lines (405, 488, 561, and 640), and 4 PMT detectors with 2 GaAsP detectors. The software capabilities include advanced 2D tracking of the cells or organelles, 3D measurement analysis, deconvolution, and tile scanning.
The LEICA SP8 confocal microscope: The microscope is built off a DMi8 inverted research microscope and comes equipped with a white light laser scan head that is tunable within the range of 470-670nm with up to eight laser lines; a 405nm laser; a filter-free spectral detector for up to five individually regulatable channels.
NIKON SMZ1270 Stereomicroscope: The lab has two sets of stereomicroscopes equipped with a camera and computer for high-speed imaging.
OTHER
Mechanical stretcher: Lab is equipped with a state-of-the-art custom-made radial mechanical stretcher controlled by the software and can stretch and compress the tissue. It also enables live imaging of cellular events like the apical expansion of MCCs and centriole amplification using a confocal microscope while applying mechanical forces.
Microinjection room: Four sets of microinjection stations that include picospritzers, micromanipulators, needle holders, and stereomicroscopes.
TECHNIQUES
Genome manipulation: CRISPR-Cas9 and morpholino oligo in frog embryos, skin organoids, and cell culture
Imaging: Confocal and Super-resolution microscopy, live imaging, Scanning Electron Microscopy (SEM), Transmission electron microscopy (TEM), and Electron Tomography (ET)
Cell culture: Generation of stem cell explants and organoids
Transcriptomics: Single cell and single nuclei sequencing
Computational biology: Mathematical modeling and machine learning
Mechanobiology: Biomechanical manipulations of cells and skin organoids
UVA FACILITIES
The Keck Center for Cellular Imaging
AFFILIATIONS
DEVELOPMENTAL GENOMICS CENTER
The Developmental Genomics Center at UVA will bridge developmental biologists with genomic and clinical translational scientists across grounds and with nearby Inova Health System and the NIH NICHD. The Center aims to integrate genomic technologies and next-generation sequencing datasets from human and animal model systems to address cutting-edge research questions in cell and developmental biology.
CENTER FOR MEMBRANE AND CELL PHYSIOLOGY
UVA’s Center for Membrane and Cell Physiology strives to understand fundamental biological processes at the highest possible spatial and temporal resolution. Our ultimate goal is to use high-end imaging, structural, biophysical, and biological and chemical probe technologies to make impactful discoveries on understanding the causes, development, and cures of diseases ranging from cardiovascular to cancer to neurological and infectious diseases.
CHILD HEALTH RESEARCH CENTER
The center's mission is to support scientists engaged in basic and clinical research to discover innovative therapies for childhood diseases. We bridge the gap between the laboratory and the bedside with cutting-edge research that improves the lives of children.
GRADUATE PROGRAMS AND TRAINING GRANTS
The Kulkarni lab is affiliated with postdoctoral and graduate T-32 training grants supported by the National Institutes of Health at the University of Virginia. Post-doctoral and Ph.D. applicants who are interested in the Kulkarni lab may be good candidates for applying for financial support from these training grants.

CONTACT
Kulkarni lab
Department of Cell Biology
Department of Biology,
University of Virginia
Charlottesville, VA 22908
Email: sk4xq@virginia.edu
Phone No: +14342976833
